Meta publishes database by AI 'ESM-2' that predicted over 600 million protein structures, prediction speed is 60 times faster than AlphaFold
Meta, which operates Facebook and Instagram, has released the database '
Announcing the ESM Metagenomic Atlas — the first comprehensive view of the 'dark matter' of the protein universe. Made possible by ESMFold, a new breakthrough model for protein folding from Meta AI.
—Meta AI (@MetaAI) November 1, 2022
More in our new blog ➡️ https://t.co/LsUhSjzqCf
1/3 pic.twitter.com/5lq48rPv5A
Explore - ESM Metagenomic Atlas
https://esmatlas.com/
Evolutionary-scale prediction of atomic level protein structure with a language model | bioRxiv
https://www.biorxiv.org/content/10.1101/2022.07.20.500902v2
ESM Metagenomic Atlas: The first view of the 'dark matter' of the protein universe
https://ai.facebook.com/blog/protein-folding-esmfold-metagenomics/
AlphaFold's new rival? Meta AI predicts shape of 600 million proteins
https://www.nature.com/articles/d41586-022-03539-1
Meta's newest AI determines proper protein folds 60 times faster | Engadget
https://www.engadget.com/metas-newest-ai-figures-out-proper-protein-folds-60-times-faster-150006068.html
Understanding the proteins that make up living organisms is extremely important in biological and medical research. However, predicting the three-dimensional structure of a protein in which a polypeptide chain in which amino acids are linked is called the `` folding problem ,'' and it is a difficult task for researchers.
In recent years, attempts have been made to predict the three-dimensional structure of proteins using AI as a method to address the protein folding problem. The protein structure analysis AI called ' AlphaFold ' developed by DeepMind, an AI research institute under the umbrella of Alphabet, has made it possible to analyze the three-dimensional structure of proteins with the same accuracy as experimental methods in a short time and at a low cost. . AlphaFold will be open sourced in July 2021 and is said to have changed the world of biology.
How is the AI `` AlphaFold '' that predicts the three-dimensional structure of proteins changing the world of biology? -GIGAZINE

In July 2022, the three-dimensional structures of more than 200 million proteins predicted by AlphaFold were released as a searchable database.
DeepMind will publish a database that allows you to easily search for over 200 million protein three-dimensional structures like Google search - GIGAZINE
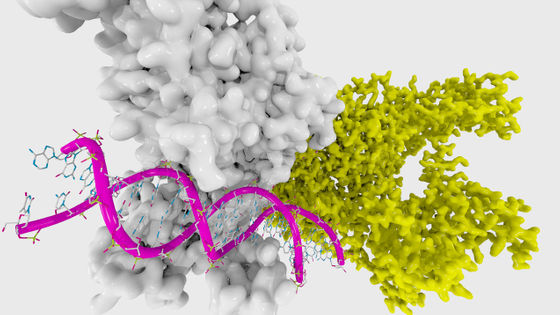
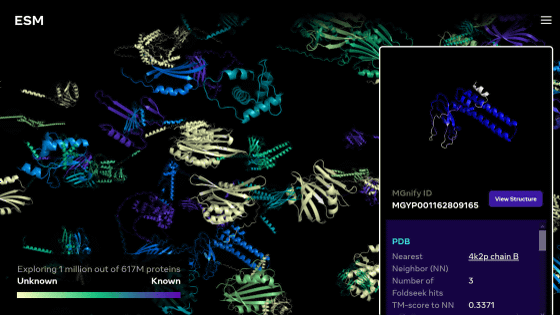
Then, in November 2022, Meta's AI research team, which operates Facebook and others, released the ESM Metagenomic Atlas, a database that predicts more than 617 million types of metagenomic protein structures. Metagenomics is a research field that deals with genomes collected directly from environmental samples, and Meta predicted the protein structure of the genome contained in the public resource ` ` MGnify90 '' that catalogs metagenomic sequences.
The research team said, 'To our knowledge, the ESM Metagenomic Atlas is the largest collection of high-resolution predicted protein structures. This database is three times larger than any existing protein structural database and comprehensively covers metagenomic proteins.' These protein structures provide an unprecedented view and new scientific insights into the breadth and diversity of the natural world and are useful in medicine, natural chemistry, environmental applications and renewables. It has the potential to accelerate the discovery of proteins that can be put to practical use in fields such as energy.'
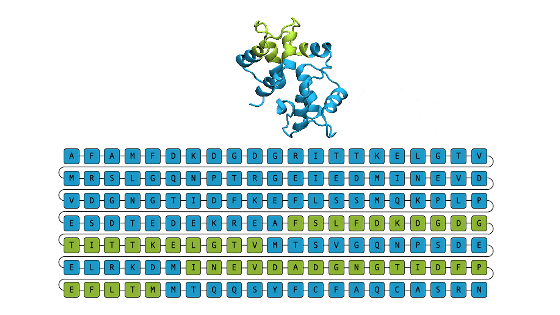
Meta's protein structure prediction AI is named 'ESMFold', and it is a model that analyzes the atoms and molecules that make up proteins as languages and predicts the three-dimensional structure from the learning data. The research team scaled up this model and developed 'ESM-2' with 15 billion parameters. ESM-2 is the largest 'protein language model' to date, predicting over 600 million protein structures in the ESM Metagenomic Atlas in just two weeks using about 2000 GPUs. reported.
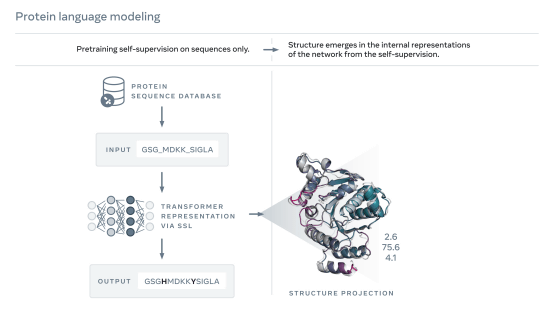
According to the research team, ESM-2's prediction accuracy is not as good as AlphaFold's, but the speed of structural prediction reaches 60 times. ``What this means is that structure prediction can be extended to much larger databases,'' said Alexander Rives, leader of the protein research team at Meta AI.
“(The metagenomic database) should cover much of the protein world that has never been seen before,” said Martin Steinegger, a computational biologist at Seoul National University. ' said.
Related Posts: