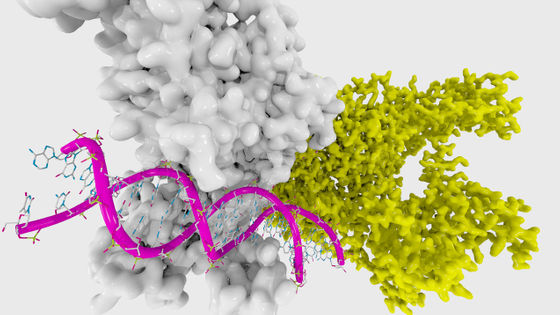
DeepMind, an artificial intelligence enterprise, develops 'AlphaFold' to predict protein three-dimensional structure
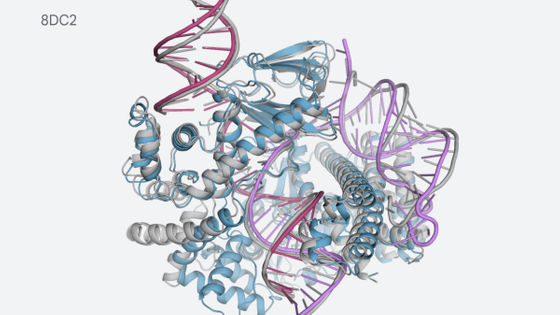
DeepMind , a brother company of Google and researching artificial intelligence, has developed a technology " AlphaFold " that predicts protein three-dimensional structure from gene sequence information. Understanding the exact three-dimensional structure of the protein is thought to bring great development to new drug development of Alzheimer's disease and Parkinson's disease .
AlphaFold: Using AI for scientific discovery | DeepMind
https://deepmind.com/blog/alphafold/

Proteins are related to almost all functions of the human body, such as contracting muscles, sensing light, converting food to energy, and so on. Protein is a macromolecular compound in which a large number of L-amino acids existing in 20 chains are linked, but how amino acid units called amino acid residues are connected to each other can be confirmed only by one- It is not mentioned. For this reason, predicting the three-dimensional structure of a protein is a problem called "protein folding problem". It is said that the larger the protein, the more complicated and difficult the modeling becomes.
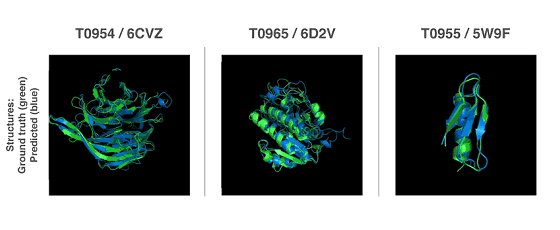
In previous studies, the three-dimensional structure of proteins was clarified by low-temperature electron microscopy , nuclear magnetic resonance , and X-ray crystallography, but both methods require trial and error for each structure, which is very costly It was. Therefore, prediction by AI attracted attention as an alternative method, but fortunately since the gene evolution technology has greatly evolved over the last few years, we were able to prepare abundant data of gene field. And DeepMind succeeded in developing AlphaFold.
Rather than using the protein folding problem solved in the past, the research team has worked out from scratch on modeling the target structure. It was found that prediction accuracy was high when predicting the material properties of the protein structure, so the research team predicted the complete protein structure using two different methods.
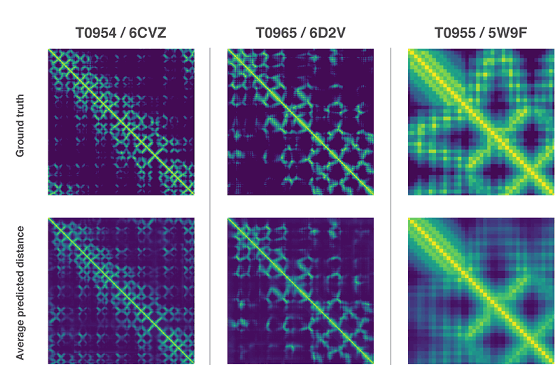
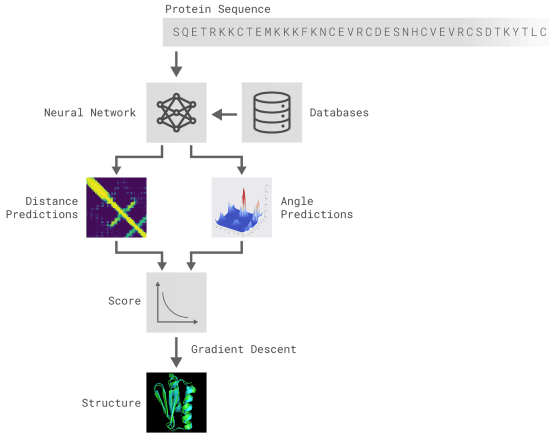
All of the above two methods use neural networks trained to predict the properties of proteins from genetic sequences. The network's predicted properties are two "distance of pair of amino acids" and "angle between chemical bonds connecting them". Based on these two predictions, it was scored how accurately the protein structure could be presented, and this score was optimized by the steepest descent method using machine learning, which enabled prediction with high accuracy Thing.
Diseases of diseases such as Alzheimer's disease, Parkinson's disease, Huntington's disease and cystic fibrosis are thought to be caused by wrong folding of proteins, predicting the structure of proteins three-dimensionally and deepening their understanding are new It is considered to be useful for the development of medicine. Also, understanding protein folding suggests that if there is progress in the field of biodegradable enzymes, the decomposition of waste such as plastics and oil may be carried out in a more environmentally-friendly manner It is.
Related Posts: