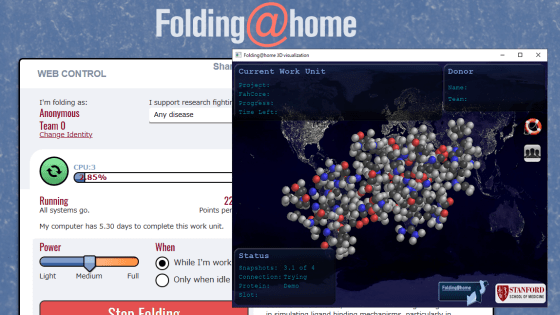
`` Folding @ home '', which can help develop new therapies for coronavirus on your own PC, starts a new small molecule screening project

Folding @ home, which performs molecular dynamics simulations of proteins by distributed computing, analyzes the protein structure of a new coronavirus (SARS-CoV-2) that is rampant around the world, and analyzes the new coronavirus infection (COVID). -19) We are working on a project to promote the development of treatment methods. It has been announced that a new small molecule screening simulation has been added to Folding @ home's analysis project.
New COVID-19 small molecule screening simulations are running on full Folding @ home! – Folding @ home
https://foldingathome.org/2020/03/30/covid-19-free-energy-calculations/
@foldingathome has launched projects to screen for small molecules that target the # COVID19 main protease.The results will help prioritize compounds for experimental testing by our collaborators.Read more at https://t.co/4wD5TSibTv @voelzlab @jchodera
— Greg Bowman (@drGregBowman) March 31, 2020
Folding @ home's project aims to rapidly develop new treatments for coronavirus infections by recruiting unused computing resources of CPUs and GPUs of PCs around the world, operating distributed computing, Analyzing the structure of the protein possessed by coronavirus. It has also been reported that Folding @ home has already outperformed the world's fastest supercomputer, Summit , as a result of people around the world providing their own computing resources.
Report that analysis system of new type coronavirus has reached processing speed exceeding supercomputer-gigazine
And on March 30, 2020, Folding @ home reported that it had launched a new batch to perform small molecule screening. The newly launched small molecule screening simulation will help prioritize molecules that inhibit an enzyme called ' protease ' of the new coronavirus.
Because the major proteases of the new coronavirus play an important role in the replication of the virus that has entered cells, inhibition of the major proteases may reduce the growth of the virus. ` ` COVID Moonshot '', an international research group created by academic and industry scientists, has found tens of thousands of promising compound candidates that inhibit major proteases, low molecular screening Will try to evaluate these molecules.
The small molecule screening project started this time uses a technique called free energy perturbation (FEP) to calculate the affinity between potential drug molecules and viral receptors. Although this method is more expensive than other methods, it seems to have the advantage of high accuracy and physical insight.
The following article explains how to join Folding @ home and cooperate with the analysis of the new coronavirus.
`` Folding @ home '' `` BOINC '' review that can help analyze the new coronavirus using your PC's computational resources-gigazine
Related Posts:
in Web Service, Science, Posted by log1h_ik