Efforts to use extra computing power of PC for research on new coronavirus

Folding @ home takes up the fight against COVID-19 / 2019-nCoV? Folding @ home
https://foldingathome.org/2020/02/27/foldinghome-takes-up-the-fight-against-covid-19-2019-ncov/
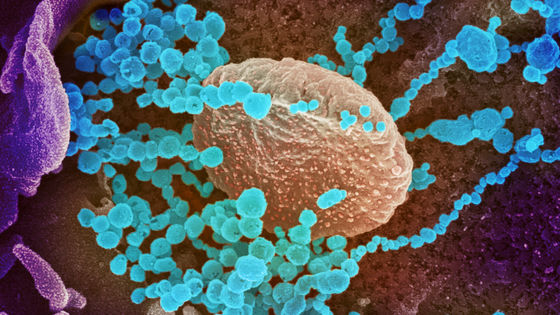
2019-nCoV is closely related to the SARS coronavirus (SARS-CoV) and is known to function similarly. Two coronaviruses enter the body by binding proteins on the virus surface to receptor proteins on lung cells. This viral protein is called a spike protein because it has protrusions (spikes), and its appearance looks like the red part in the following image.
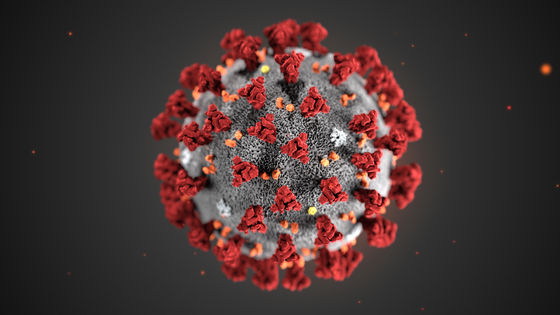
By Alissa Eckert, MS; Dan Higgins, MAM
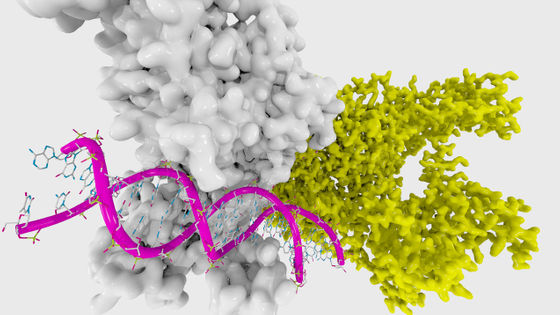
The spike protein on the virus surface of 2019-nCoV binds to the angiotensin converting enzyme 2 (ACE2) receptor expressed on the surface of lung cells and enters the human body. Therefore, therapeutic antibodies are expected to be a type of protein that can prevent spike proteins from binding to the ACE2 receptor. Although therapeutic antibodies for SARS-CoV have already been developed, developing antibodies for 2019-nCoV requires a deeper understanding of the structure of the spike protein of 2019-nCoV and the binding process with the ACE2 receptor There is.
Proteins move in small increments, taking various shapes as they fold and open. Therefore, the shape of the spike protein of 2019-nCoV is not only one. Folding @ home wrote, 'We need to study how proteins fold into other shapes so that we can better understand how spike proteins interact with the ACE2 receptor and design antibodies.' Although the structure of the spike protein of SARS-CoV has already been clarified to some extent, 2019-nCoV is a different mutation.
Based on this information, Folding @ home's goal is to model the structure of the 2019-nCoV spike protein and build a computational model to identify the target sites for therapeutic antibodies. However, because it requires a great deal of computing power, we have announced details of our efforts to seek help from users.
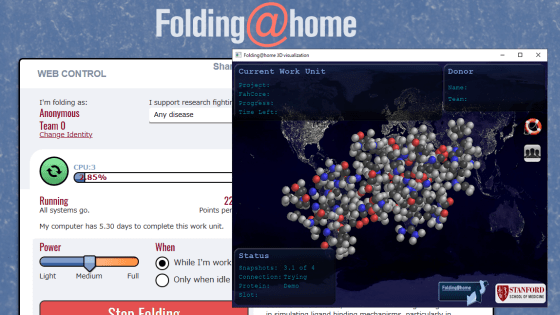
No special registration is required to cooperate with Folding @ home, and you can download and install the dedicated program distributed on the following page.
Start folding – Folding @ home
https://foldingathome.org/start-folding/
Related Posts: