Google DeepMind releases new AI model for protein design: AlphaProteo
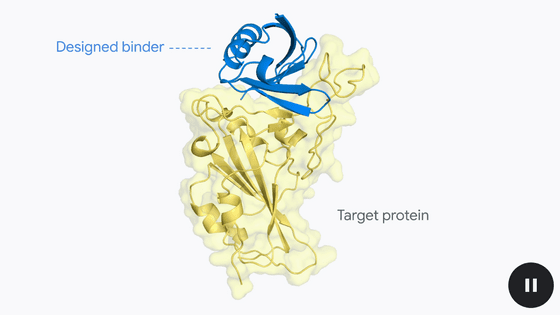
Google DeepMind has announced a new AI model called ' AlphaProteo ' for designing 'proteins that bind effectively to target proteins.'
AlphaProteo generates novel proteins for biology and health research - Google DeepMind
Like a key fitting into a lock, proteins bind to other proteins to carry out various cellular processes. While DeepMind's protein structure prediction tools, such as AlphaFold , have provided a wealth of insight into how proteins interact with each other, they have not been able to design new proteins to interact with them.
DeepMind's AlphaProteo is an AI system for designing new high-strength protein binders, which will make protein design, a process that has traditionally required a lot of experimentation, much easier. It is said to be able to speed up research in a wide range of fields, including drug development, visualization of cells and tissues, understanding and diagnosing diseases, and acquiring resistance to pests in crops.
The DeepMind team targeted seven proteins and used AlphaProteo to generate protein binders, which they say were the first AI tool to design suitable protein binders for a protein called VEGF-A, and that the candidates were more likely to actually bind to their targets than the best previous methods, with binding affinities between three and 300 times higher.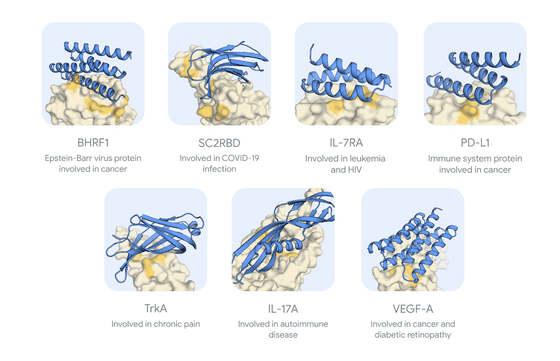
AlphaProteo was trained using vast amounts of protein data from the Protein Data Bank and over 100 million structure predictions from AlphaFold. From this data, AlphaProteo learns the various ways molecules can bind to each other, so given a target protein's structure and preferred binding site, it can generate candidate proteins that bind to the target protein at that site.

The image below is a graph showing the probability that proteins generated by AlphaProteo and conventional methods can successfully bind to the target protein. The blue data shows that AlphaProteo has a higher probability of successful binding than conventional methods, and it can be seen that the number of candidates required to find a protein that can bind successfully is reduced.
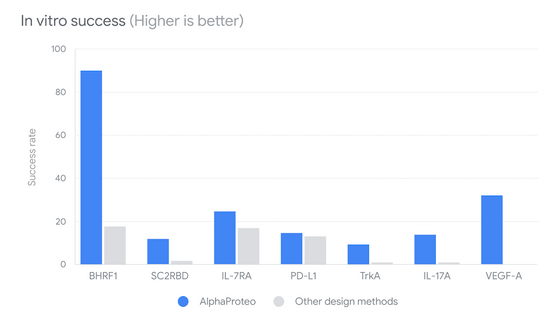
The 'affinity' score, which indicates the strength of binding, is shown in the figure below. The graph is on a logarithmic scale, and the smaller the number, the stronger the newly designed protein can bind to the target protein. AlphaProteo, shown in blue, was able to create stronger bonds with all proteins than the conventional ones.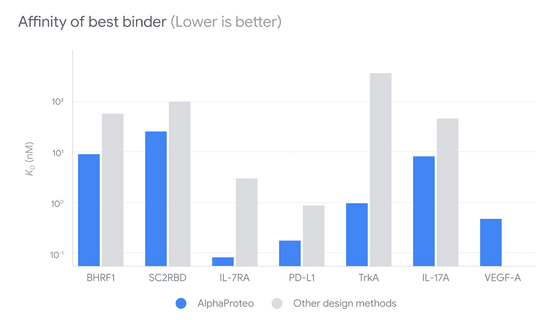
These results show that AlphaProteo can significantly reduce the time required for initial experiments to create protein binders. However, it is not possible to create binders for all proteins. Even with AlphaProteo, we were unable to design a binder for TNFɑ, a protein associated with autoimmune diseases such as rheumatoid arthritis, for which computational analysis has shown that it is difficult to design binders.
DeepMind said in the release that it will 'continue to improve and extend AlphaProteo with the goal of eventually addressing challenging targets like TNFɑ.'
Related Posts: