Using AI to design proteins that block snake venom

A research team led by Professor David Baker of the University of Washington, who won the Nobel Prize in Chemistry in 2024, announced that they have designed a new protein that can inhibit some of the toxins contained in snake venom using AI-based three-dimensional protein structure prediction.
De novo designed proteins neutralize lethal snake venom toxins | Nature
https://www.nature.com/articles/s41586-024-08393-x
Researchers use AI to design proteins that block snake venom toxins - Ars Technica
https://arstechnica.com/science/2025/01/researchers-use-ai-to-design-proteins-that-block-snake-venom-toxins/
Snake venom contains a variety of toxins, including neurotoxins that cause breathing difficulties, hemorrhagic toxins that inhibit blood clotting, and cytotoxins that cause tissue damage. Most of these toxins are proteins, so if you are bitten by a snake, the only way to treat it is to inject a serum containing an antitoxin against the venom. However, this serum needs to be kept refrigerated and has a short shelf life. Generally, most snakebite accidents occur deep in the mountains or in remote areas far from cities, so the difficulty of preserving it is a major obstacle to its operation.
If a smaller, more stable protein with the same function as this antitoxin could be found, it might be possible to use genetic engineering to produce the protein in bacteria or other organisms, creating an antitoxin that does not require refrigeration.
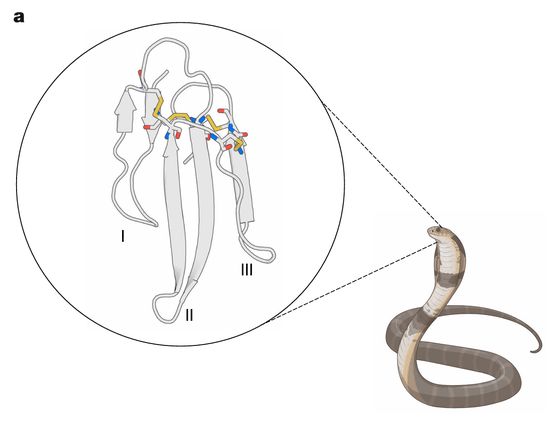
Professor Baker's research team recently announced an antivenom from a group of proteins called three-finger toxins (3FTx), found in venomous snakes of the Elapidae family. 3FTx contains neurotoxins that cause respiratory paralysis, as well as cytotoxins that cause limb deformation and skin necrosis. As the name suggests, 3FTx has a three-dimensional structure in which three beta sheets are extended. Because 3FTx has low immunogenicity , there was a problem that it was difficult to obtain a sufficient neutralizing antivenom using conventional serum treatment, and there was also a time constraint in which the effect decreased if administration was delayed.
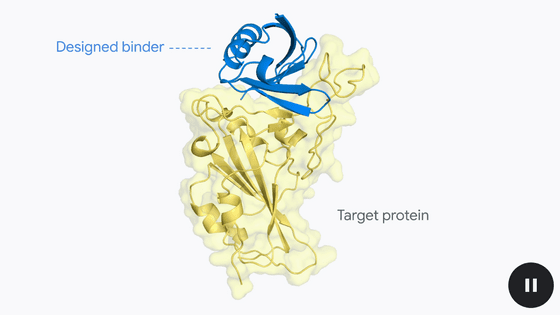
The team used a deep learning-based technique called 'RFdiffusion' to analyze the toxin's structure in detail and find amino acid chains that fit perfectly into the three beta-sheet structure. They then used an AI package called 'ProteinMPNN' to identify the amino acid sequences that had the chains analyzed by RFdiffusion. The new protein's structure was then verified using AlfaFold 2 and Rosetta, which predict protein 3D structures. After selecting the one that showed the strongest interaction with 3FTx from among 44 computer-designed proteins, the team again used RFDiffusion to search for mutants of this protein that were more likely to be effective.
As a result, they succeeded in developing three proteins named SHRT, LNG, and CYTX, which correspond to three different toxins, and which were stable even at relatively high temperatures and had the ability to bind to the toxins.
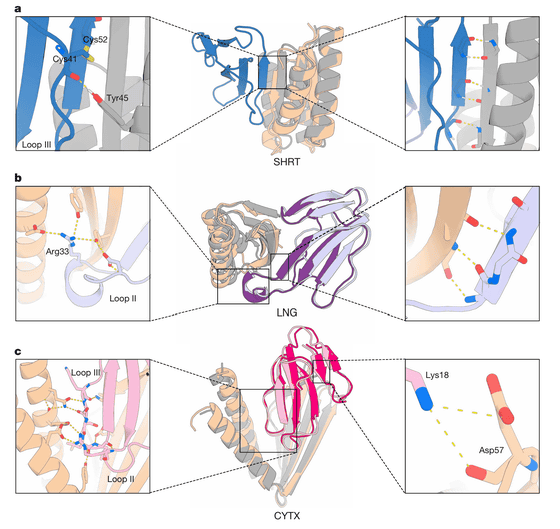
In experiments using mice, the neurotoxins SHRT and LNG were found to be significantly more effective, with survival rates of 100% for SHRT and 60% for LNG even 30 minutes after administration of a lethal dose of the toxin, and no paralysis of the limbs or breathing was observed in the surviving mice.
However, the cytotoxin CYTX did not show any significant effect. The research team stated that because the mechanism by which the cytotoxin of 3FTx destroys the cell membrane of the skin has not been fully elucidated, further research and optimization of the structure of CYTX are required to improve its effectiveness.
It is important to note that this research and experiment is still in the proof-of-concept stage, and snake venom is diverse. However, whereas conventional serum was produced using animals, the protein designed by this method can be produced using bacteria, making it possible to produce antivenom at a lower cost than conventional methods. In addition, since it is easier to store, the hurdles of transportation and procurement are greatly reduced, and it is expected that treatment costs will be reduced.
The research team argued that 'the RFdiffusion design methodology can also be applied to the development of neutralizing proteins for other medically important toxins, potentially accelerating the development of antitoxins that can be used against a wider range of snake venoms.'
Related Posts:
in Science, Posted by log1i_yk