Open source implementation of protein structure prediction AI model 'AlphaFold3' finally released

Google DeepMind's AI model for predicting the structures of proteins and other biological molecules, AlphaFold 3, has been implemented in an open source manner and the ongoing research project has been published on GitHub.
GitHub - Ligo-Biosciences/AlphaFold3: Open source implementation of AlphaFold3
Show HN: An open-source implementation of AlphaFold3 | Hacker News
https://news.ycombinator.com/item?id=41448439
AlphaFold 3 is an AI model that predicts the structure and interactions of life, which is made up of millions of combinations of molecules, and is expected to be useful in fields such as drug development and genomics research. Since its announcement, most of its functions have been available for free through a public server.
Google DeepMind announces 'AlphaFold 3', an AI model that can predict the structure and interactions of all biological molecules with extreme accuracy - GIGAZINE
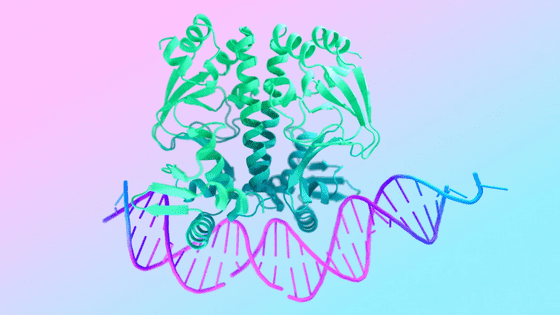
However, when AlphaFold 3 was first announced, questions were raised about its accuracy and reproducibility because the model's code was not made public.
The development team hopes to gradually open-source features of AlphaFold 3, advancing research towards a full open-source implementation that the entire community can use freely.
AlphaFold 3 is released under
The development team said, 'AlphaFold 3 has a wide range of applications, including CRISPR gene editing and predicting how drugs bind to cancer. For now, the repository's main use is research and development, but we plan to add user-facing features in the future as soon as the ligand protein and nucleic acid prediction functions are ready.'

Related Posts: