'Virtual Pharmacology' dramatically accelerates the existing drug discovery process and makes it possible to identify unknown drugs

by freestocks.org
The scientists at the University of California, San Francisco, in collaboration with researchers at the University of North Carolina, have developed the world's largest virtual pharmacology platform, and have found that the platform can be used to identify very powerful new drugs. This virtual pharmacology platform contains more than 100 million virtual molecules that have never been found in nature or synthesized before, not only dramatically changing the existing drug discovery process It has been suggested that it could have a major impact on the pharmaceutical industry.
Ultra-large library docking for discovering new chemotypes | Nature
https://www.nature.com/articles/s41586-019-0917-9
'Virtual Pharmacology' Advance Tackles Universe of Unknown Drugs | UC San Francisco
https://www.ucsf.edu/news/2019/02/413236/virtual-pharmacology-advance-tackles-universe-unknown-drugs
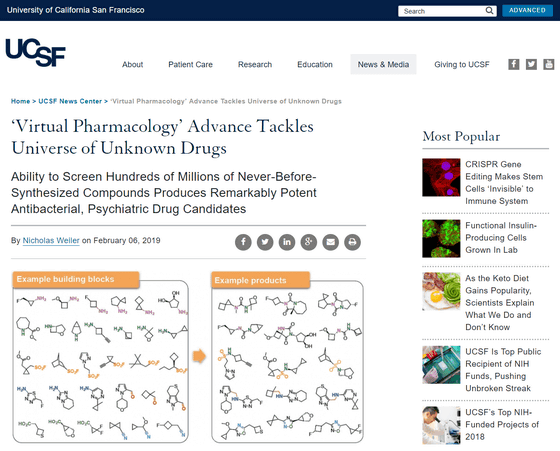
With the rapid development of “ atomic scale molecular imaging ” technology and “ virtual pharmacology ” pioneered by the University of California San Francisco, the chemical structures of important biological targets are elucidated, and millions of drug molecules (drug treatment (drug treatment) It is becoming possible to quickly simulate how molecules (which can be applied) can bind to a target. This will allow us to identify effective drug candidates without physical synthesis or testing.
Researchers estimate that the number of potential molecules will be close to the number of observable atoms in the universe, but the number of drug molecules contained in existing drug discovery databases is at best There are several million kinds. Therefore, by using virtual pharmacology and molecular imaging technology, using a much larger number of drug molecules than existing drug discovery databases, by creating a platform that can identify much more abundant new drugs than conventional methods. May dramatically evolve the drug discovery process.
Brian Shoychett, a professor of pharmaceutical chemistry at the University of California, San Francisco, who noticed such an idea, and John Irwin, a part-time associate associate professor, collaborated with the Ukrainian chemical supplier "Enamine" to work on virtual medicines. Using his knowledge of science and molecular imaging technology, he has developed a "virtual pharmacology platform" and published it in the scientific journal Nature.
by Simone van der Koelen
Enamine is a company that has generated many drug compounds that have not existed until now at a cost of about $ 100 (about 11,000 yen) per molecule over the past 10 years. By combining tens of thousands of standard chemical constituents with one another using hundreds of chemical reactions, we are building an efficient method to produce more than 100 million types of drug compounds on demand .
Shoichett and Erwin have combined the data on the vast number of virtual drug compounds that they obtained with Enamine in a free drug discovery database called ZINC . ZINC contains more than 750 million hypothetical drug compound data at the time of writing, and as Enamine and other suppliers add new data daily, the number will increase over time It is estimated to be.
The research team uses a technique called " docking " to transform Enamine's hypothetical drug compound data into a three-dimensional chemical model that is compatible with computational pharmacological approaches. As a result, it is possible to quickly simulate in 3D how the "hundreds of kinds of virtual drug data" and "specific biological targets" contained in the database will be combined.

by Ani Kolleshi
Based on the exponential growth rates so far, ZINC is predicted to include more than 100 million types of 3D models that have not been synthesized by 2020. The research team says "Our platform is currently capable of screening 100 times more molecules than those available in common drug screening libraries. Molecular diversity will continue to increase, and more than 1,000 times more molecules soon. You will be able to screen. "
In order to prove the power of the virtual pharmacology platform that the research team has built, two unrelated biological targets in the ZINC database (related to β-lactamase related to antibiotic resistance, psychosis and addictive behavior) Search for drug molecules that are effective against dopamine D4 receptors. Using a virtual pharmacology platform, after screening hundreds of millions of new drug candidates, we succeeded in detecting hundreds of new drugs that have never been synthesized. These included drugs known to be the most effective as existing beta-lactamase inhibitors, and one of the most potent dopamine receptor activators.
"The D4 binding compound identified from ZINC is one of the most powerful ever reported," said Michael Hooker, Ph.D. Professor of Medicine at the University of North Carolina School of Medicine. Screening is a big step towards creating more effective medicines for many diseases. "
By using ZINC, it is possible to test whether even previously synthesized drug molecules act on a specific substance, so researchers should identify more potent drug candidates. It is said that you will be able to
Related Posts:
in Science, Posted by logu_ii