Google DeepMind open-sources 'AlphaFold 3,' a system that can predict the structure and interactions of all biological molecules using AI, to accelerate scientific discovery and drug discovery
AlphaFold , an AI developed by Google DeepMind in 2018 to predict the three-dimensional structure of proteins from amino acid sequence information, was open-sourced in 2021 with the aim of accelerating research in many important fields. In May 2024, the company released AlphaFold 3 , an AI model that can predict the structures and interactions of more biological molecules. Although the complete model was not initially released, AlphaFold 3 was also open-sourced, following the previous model.
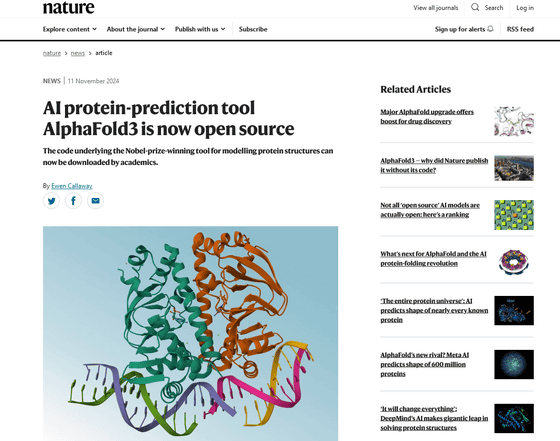
AI protein-prediction tool AlphaFold3 is now open source
https://www.nature.com/articles/d41586-024-03708-4
Proteins are substances involved in almost all biological processes, such as muscle contraction, blood transport, light sensing, and food energy conversion. However, most of the more than 200 million proteins discovered by humans have such complex structures that only their amino acid sequences are known. Predicting the three-dimensional structure from the amino acid sequence is called the ' protein folding problem ,' and has been a major problem in biology for many years. Google DeepMind's AlphaFold has used AI to greatly improve the three-dimensional structure analysis of proteins, which was previously too time-consuming and costly.
How is the protein structure predicting AI 'AlphaFold' changing the world of biology? - GIGAZINE

After the release of AlphaFold in 2018, AlphaFold 2 was released in 2020, which made fundamental advances in protein structure prediction. AlphaFold 3, announced in May 2024, will be able to handle a wide range of biological molecules in addition to proteins, and is expected to boost scientific innovation in a wide range of fields, from research into renewable materials and resilient crops to drug design and genomics research.
On November 11, 2024, Google DeepMind will open source AlphaFold 3, following its predecessor. The software code for AlphaFold 3 will be published on GitHub and available for anyone to download and use.
John M. Jumper, a senior research scientist who won the 2024 Nobel Prize in Chemistry with Demis Hassabis, founder of Google DeepMind, said, 'I'm really looking forward to seeing what people do with this' when AlphaFold 3 is open-sourced. In addition, Google DeepMind computer scientist Pushmeet Kohli posted on X, 'The AlphaFold 3 model code and weights are now available for academic purposes. We at Google DeepMind are looking forward to seeing how the research community will use AlphaFold to tackle open problems and new areas of research in biology.'
The #AlphaFold 3 model code and weights are now available for academic use. We @GoogleDeepMind are excited to see how the research community continues to use AlphaFold to address open questions in biology and new lines of research. https://t.co/kVB9hWJZTI https://t.co/k0Fz85OQzB
— Pushmeet Kohli (@pushmeet) November 11, 2024
Google DeepMind initially made AlphaFold 3 available only via a web server in order to strike the right balance between enabling access for research purposes and protecting commercial interests. However, it was strongly criticized by scientists for violating the openness and peer review standards set by Nature magazine, which published the paper, and for releasing AlphaFold 3 without the code, model weights, and parameters obtained by training the software on protein structures and other data, which 'impaired the reproducibility of structural analysis.' So Google DeepMind changed its policy and provided an open source version of AlphaFold 3 about six months after its release.
Anyone can download AlphaFold 3 from GitHub, but at the time of writing, scientists affiliated with academic institutions must request access to the training weights required for stable structure analysis. GitHub states, 'To request access to the AlphaFold 3 model parameters, please submit a form. Access will be granted at Google DeepMind's sole discretion.'
Related Posts: