AI succeeds in discovering genes related to autism from a region called 'junk DNA'

by
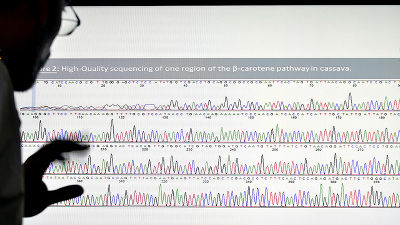
Proteins that make up the organism's body are synthesized based on DNA sequences, but only 3% of human genomes are known to be directly involved in protein synthesis, the remaining 97% It is said that ' Junk DNA ' does not know what functions it has. However, using deep learning to analyze the human genome, it has been reported that it was possible to newly discover a gene involved in autism from the region that has been considered junk until now. Published in Nature Genetics.
Whole-genome deep-learning analysis identifications contribution of noncoding mutations to autism risk | Nature Genetics
https://www.nature.com/articles/s 41588-019-0420-0
New Causes of Autism Found in 'Junk' DNA
https://www.simonsfoundation.org/2019/05/27/autism-noncoding-mutations/
The research group that discovered autism-related genes from junk DNA by deep learning analysis is the research group of Jian Zhou et al. The research group analyzed the whole genome of 1790 autism patients and their families with deep learning. In addition, in order to exclude the influence of 'Autism gene inherited from relatives' from the analysis results, the genome of the autism patients whose autism patients are in close relatives is excluded from the genome to be analyzed. I am doing

by
As a result, they have succeeded in finding gene sequences related to autism from junk DNA not directly involved in protein synthesis. The analysis also detected genes that synthesize proteins that have been known to be associated with autism, but their gene sequences are only about 30% of the entire gene sequences identified this time, and the rest are Seventy percent were found in the region of junk DNA.
Until now it has been speculated that junk DNA 'does not have any regulatory function without synthesizing a protein,' but no specific case has been identified. Because junk DNA is just a random base sequence at first glance with no context at first glance, comparing the genome of a patient with the genome of a non-patient does not know what that difference means, as the cause It can not be identified.

by bialasiewicz
With deep learning, even if the behavior of the gene sequence is not known, it is possible to focus on the difference in the sequence itself to predict the association with the disease. This will enable us to focus on mutations in genes that are closely related to disease, rather than randomly comparing genomes.
Olga Troyanskaya, a member of the research group at Stanford University, analyzes the findings using “deep learning to identify causes of various diseases not only autism but of which the causative gene is not known. It will be possible, ”said the view that 97% of the genes that have not been considered until now can be expected to find many.
Related Posts:
in Science, Posted by log1l_ks