DeepMind publishes a database that allows you to easily search over 200 million protein 3D structures as easily as Google search
Google's parent company Alphabet's AI laboratory 'DeepMind' is developing a number of AI such as protein structure analysis AI '
AlphaFold reveals the structure of the protein universe
https://www.deepmind.com/blog/alphafold-reveals-the-structure-of-the-protein-universe
Proteins are the main constituents of living organisms, and identifying the structure of proteins greatly contributes to the development of a wide range of fields such as biology and drug development. Amino acids bind to each other to form a polypeptide chain, and the polypeptide chain is folded to form a three-dimensional structure. Although the amino acid sequence can be analyzed relatively easily, the analysis of the three-dimensional structure after the polypeptide chain is folded is called the ' folding problem ' and has been regarded as a major obstacle to protein-related research due to the difficulty of work. ..
Techniques such as cryo-electron microscopy and X-ray crystallography have been developed to solve the folding problem, but these techniques required a lot of time and money. The protein structure analysis AI 'AlphaFold' developed by DeepMind in 2018 can predict the three-dimensional structure of proteins with the same accuracy as cryo-electron microscopy and X-ray crystal structure analysis, and researchers can use AlphaFold for a short time. Moreover, it has become possible to analyze the protein structure at low cost. In addition, AlphaFold was made open source in July 2021 and was used by more than 500,000 researchers in the year until July 2022, and many research results on plastic pollution and drug resistance were announced. I am.
How is the AI 'AlphaFold' that predicts the three-dimensional structure of proteins changing the world of biology? --GIGAZINE

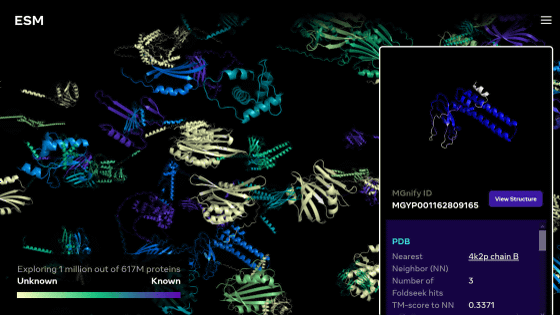
DeepMind had released the protein structure data elucidated by AlphaFold as AlphaFold DB, but in partnership with the European Institute of Molecular Biology, more than 200 million protein structures can be viewed on AlphaFold DB. Announced that it has become. There are a wide variety of protein structures that can be viewed, such as animals, organisms, bacteria, and fungi, and it is possible to find the desired protein structure information with as simple an operation as Google search.
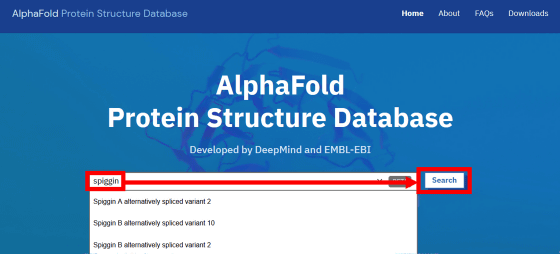
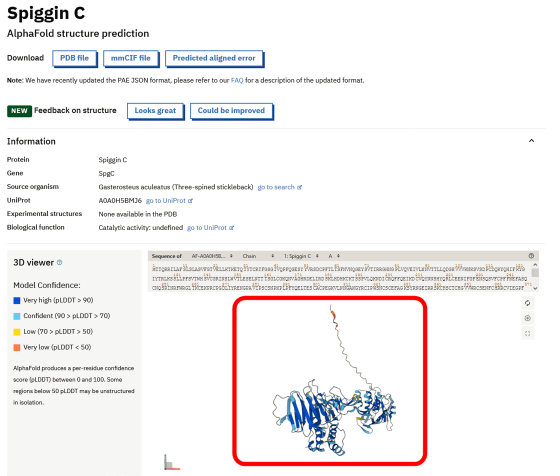
The actual AlphaFold DB screen is below. There is a search box on the top page of AlphaFold DB, enter the name of the protein you want to search and click 'Search' ...
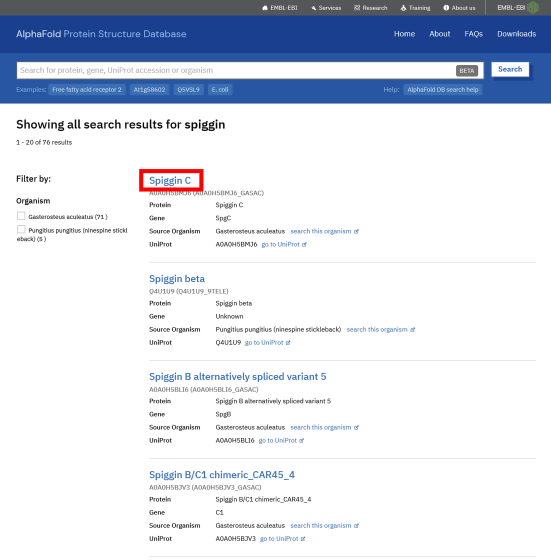
Like this, the proteins related to the entered name are displayed in a row. If you click on the protein you are interested in from the search results ...
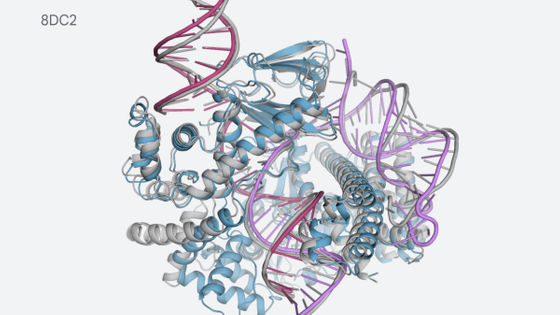
The three-dimensional structure is displayed along with the protein name and database serial number. The displayed three-dimensional structure can be moved with the mouse.
Related Posts:
in Web Service, Web Application, Science, Posted by log1o_hf