Artificial intelligence company DeepMind releases protein structure analysis algorithm 'AlphaFold' as open source, making it available to anyone
by OIST
DeepMind, an artificial intelligence company affiliated with Google, announced that it has open sourced the algorithm 'AlphaFold ' for predicting and analyzing the three-dimensional structure of proteins, which has been under development for some time, and released the source code to GitHub free of charge. .. 'The open source of AlphaFold allows users to elucidate the three-dimensional structure of proteins with unprecedented accuracy,' DeepMind said.
AlphaFold Open Source | DeepMind
https://deepmind.com/research/open-source/alphafold
GitHub --deepmind / alphafold: Open source code for AlphaFold.
https://github.com/deepmind/alphafold
Proteins that perform various functions in the human body are macromolecular compounds in which 20 types of amino acids decoded from the base information (genome) that make up genes are bound. However, only the primary sequence information such as 'which amino acids are connected' can be understood from the base information. In order to analyze how a protein functions in the body, it is necessary to know the three-dimensional structure of the protein. However, the three-dimensional structure cannot be known only from the amino acid sequence information. This problem of 'how to elucidate the three-dimensional structure of proteins ' is called the 'folding problem ' and has been tackled by many scientists.
The elucidation of the three-dimensional structure itself has already been carried out, and in the past, the three-dimensional structure of proteins was specified by nuclear magnetic resonance , cryo-electron microscopy , X-ray diffraction, etc., but all of them are expensive. However, there was a problem that it would be difficult unless the research team and medical institution had abundant budget. In addition, a lot of computing power is required to elucidate the actual three-dimensional structure from the data, and it is necessary to perform arithmetic analysis with distributed computing such as supercomputers and Folding @ home , so not only the cost of funds but also the cost of time. It also took.
Developed by DeepMind in 2018, AlphaFold is a technology for predicting the three-dimensional structure of proteins from gene sequence information using machine learning. This makes it possible to elucidate the three-dimensional structure of proteins without incurring conventional costs.
Artificial intelligence company DeepMind develops 'AlphaFold' that predicts the three-dimensional structure of proteins --GIGAZINE
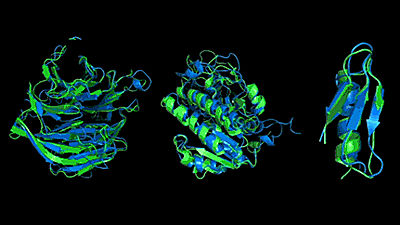
In fact, when AlphaFold was evaluated in a protein structure prediction contest, it was reported that it recorded higher accuracy than other computer programs.
'AlphaFold' developed by DeepMind has shown a path with the power of AI to the super-difficult problem of biology from 50 years ago, and research is accelerating --GIGAZINE

And on July 16, 2021, DeepMind co-founder Demis Hassabis announced that AlphaFold version 2.0 was open sourced and the source code was published on GitHub.
Last year we presented #AlphaFold v2 which predicts 3D structures of proteins down to atomic accuracy. Today we're proud to share the methods in @Nature w / open source code. Excited to see the research this enables. More very soon! Https: //t.co/6uiV51Xly5 https://t.co/CLo7EKubBT pic.twitter.com/5dAgg9mOMN
— Demis Hassabis (@demishassabis) July 15, 2021
At the same time, a paper about AlphaFold was published in the academic journal Nature.
Highly accurate protein structure prediction with AlphaFold | Nature
https://doi.org/10.1038/s41586-021-03819-2
Following the open source of AlphaFold, DeepMind's parent company, Google CEO Sundar Pichai, said, 'Today, we are pleased to announce DeepMind's groundbreaking method called AlphaFold, along with open source code. Is an exciting step for the scientific community to accelerate research in many important areas. '
Great to see the methods for @DeepMind's AlphaFold breakthrough published today along with the open-source code. This is an exciting step forward that will help enable the scientific community to accelerate research in many important areas. Https://t.co / 1yAgo1lVJ9
— Sundar Pichai (@sundarpichai) July 15, 2021
Related Posts: